Table of contents
Introduction
This TechNote explains how to solve and fit differential equations with pro Fit. First we discuss simple differential equations, then coupled differential equations. Both types of equations are illustrated by an example.
The example files used in this technote can also be downloaded as a zipped file.
Simple differential equations
Simple differential equations are of the general type:
dy/dt = f(x, y, a1, a2,… aN) (1)
where a1, a2,… aN are the parameters to be fitted.
To fit such an equation to a data set {y(xi), xi}, you must integrate it. Sometimes, you can carry out this integration analytically, which is the easiest and fastest way for fitting. Often, however, you will not be able find an analytical integral of Eq. (1) and you must resort to the methods described here.
Example 1 – Alcohol
To discuss a solution of a differential equation of the type of Eq. (1), let us look at a simple problem. Imagine you are a medical student running an experiment on a friend of yours. This experiment involves the following steps:
- At an arbitrary starting time (let’s call it time 0), you inject some alcohol into a vein of your friend.
- After having done so, you take blood samples from your friend and determine the alcohol concentration and observe how this concentration decreases.
This is a plot of your measurements:
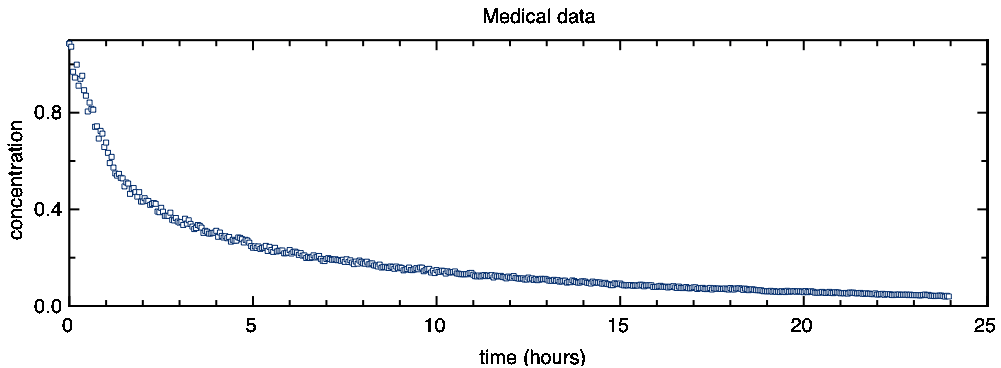
In nature, such decays are often ruled by a simple exponential equation, i.e. they are of the type
y = a exp(-t/t0) (2)
where y is the alcohol concentration, t the time, a is the concentration at time 0 and t0 the decay rate. To test if a data set follows such a function, you can plot it with logarithmic y-axis. In such a plot, the data points should appear on a straight line. The following is a logarithmic plot of your data set:
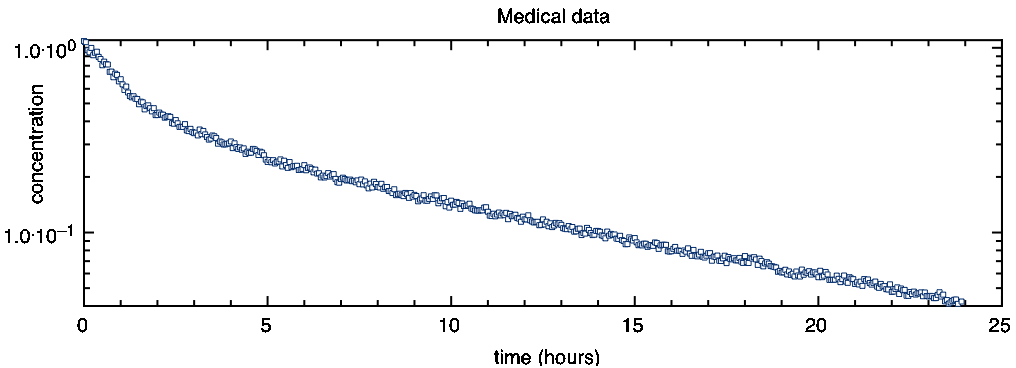
As you can see, this data does not decay with a simple exponential. At the beginning it is decaying faster than at the end.
There are various other models that might fit your data, such as a sum of two exponential decays. However, for the present discussion, let us look at a non-standard model, the one you would like to write a thesis about.
Your thoughts are the following: The exponential decay model, Eq. (2), comes from the assumption that the rate of alcohol absorption is proportional to the current concentration of alcohol in the blood. Since the rate of absorption corresponds to the negative value of the change dy of concentration in a time interval dt, we have
dy/dt = -k·y (3)
where k is the decay constant. Equation (3) is the classical theory that, when integrated, leads to the solution of Eq. (2).
As we see from the logarithmic plot, Eq. (3) cannot be a good model. So you are wondering: “What if the absorption rate dy/dt is not linearly dependent on the current concentration y?” To refine Eq. (3), you therefore want to add higher powers of y on its right side. For a first approximation, you want to take into account all powers of y up to 3, i.e. you want to test if your data could be explained with the model
dy/dt = k1·y + k2·y2 + k3·y3 (4)
Where k1, k2, and k3 are the decay rates (can be negative).
In an ideal world, you are an excellent mathematician and therefore able to find an analytical solution of Eq. (4). However, we are not living in an ideal world, and this paper is about fitting differential equations, so we will not discuss analytical solutions.
To fit Eq. (4) in pro Fit, you need to write a function that calculates the integral of Eq. (4) numerically. There are various ways to do this, some of them simple, others sophisticated. Let’s have a look at a few of them.
Simple solution:
The easiest way to integrate Eq. (4) is by converting it into a “difference equation” of the type:
y(t+Dt) = y(t) + Dt (k1y + k2 y2 + k3 y3) (5)
Eq. (5) is true if Dt is small enough. Therefore, to numerically integrate Eq. (4), you can use Eq. (5) to calculate, from y(0) at time 0, y at later times Dt, 2Dt, 3Dt, etc. Unfortunately, we don’t know y(0). To overcome this problem, we make y(0) a parameter that we will evaluate during fitting.
The following function implements this numerical integration of Eq. (5):
function NonlinearDecay;
const aStep = 0.01;
var t, dt;
begin
y := a[4]; {start value}
t := 0; {start time}
dt := aStep; {step width}
while t < x do
begin
if t+dt > x then dt := x-t;
y := y + dt*(y*(a[1] + y*(a[2] + y*a[3])));
t := t+dt;
end;
end;
Note that a[1] is k1, a[2] is k2, a[3] is k3, a[4] is y(0), and x is t. The while-loop calculates y(t) starting from y(0) by running through Eq. (5) again and again using the step width given in the constant “aStep”.
To fit the above function, you should first enter rough approximations for a[1] (e.g. -0.1) and a[4] (e.g. 1) . Then choose “Nonlinear Fit…” from pro Fit’s Calc menu. You will see that our function fits the data well:
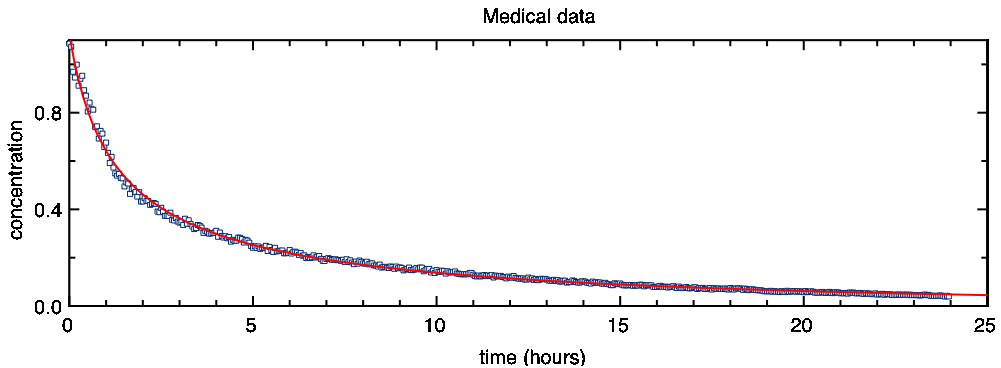
The integration used by the function NonlinearDecay is rather slow. Fitting our data took around 12 seconds on a moderately new mac. This is an acceptable time in practice, but fitting a larger number of data points or a more complicated function can be quite boring. There are various ways for making the fitting of differential equations faster. Some of them are described in the following:
Faster solution 1: using a better integration algorithm
There are several methods for efficient integration of differential equations. They are described in the standard text books (see e.g. Numerical Recipes in Pascal or Numerical Recipes in C by Press, Flannery, Teukolsky, Vetterling, Cambridge University Press). In the following, we will discuss one of them, the Runge-Kutta method.
This method is based on a fairly simple theory of Taylor series. It allows to calculate y(t+Dt) from y(t) and Dt using much larger step widths Dt than Eq. (5). It uses the following:
y(t+Dt) = Dt (m1 + 2 m2 + 2 m3 + m4)/6 (6)withm1 = f(t, y(t))
m2 = f(t + Dt/2, y(t + Dt/2 m1))
m3 = f(t + Dt/2, y(t + Dt/2 m2))
m4 = f(t + Dt, y(t + Dt m3))
where f was introduced in Eq. (1).
The following is an implementation of our program using the Runge-Kutta method:
function NonlinearDecay2; {Runge-Kutta}
const aStep = 0.05;
var t, dt, m1, m2, m3, m4, y1;
begin
y := a[4]; {start value}
t := 0; {start time}
dt := aStep; {step width}
while t < x do
begin
if t+dt > x then dt := x-t;
m1 := y*(a[1] + y*(a[2] + y*a[3]));
y1 := y + dt*m1*0.5;
m2 := y1*(a[1] + y1*(a[2] + y1*a[3]));
y1 := y + dt*m2*0.5;
m3 := y1*(a[1] + y1*(a[2] + y1*a[3]));
y1 := y + dt*m3;
m4 := y1*(a[1] + y1*(a[2] + y1*a[3]));
y := y + dt*(m1 + 2*m2 + 2*m3 + m4)/6;
t := t+dt;
end;
end;
This is twice as fast with comparable accuracy and fits our data in approximately 8 seconds.
Faster solution 2: using the last result
Another approach for speeding up the function is the following: Whenever our function calculates a value y(t1), it stores y and t1 and, when it is called the next time with x = t2 and t2 > t1, integration starts at t1 with y(t1) instead of starting at t = 0 with y(0).
This method has one drawback: Whenever the function’s parameters change, the value y(t1) from the previous call (which was calculated for other parameters) becomes invalid. Therefore we must use the procedure First to reset y(t1). This is shown in the following:
function NonlinearDecay3;
const aStep = 0.01;
var t, dt;
var lastX, lastY;
procedure First; {called when lastX, lastY are stale}
begin
lastX := 0;
lastY := a[4];
end;
begin
if x >= lastX then begin
y := lastY; {start value}
t := lastX; {start time}
end
else begin
y := a[4]; {start value}
t := 0; {start time}
end;
dt := aStep; {step width}
while t < x do
begin
if t+dt > x then dt := x-t;
y := y + dt*(y*(a[1] + y*(a[2] + y*a[3])));
t := t+dt;
end;
lastX := x;
lastY := y;
end;
For simplicity, this function does not use the Runge-Kutta method.
In its present form, it fits our data in 10 seconds. This is not much faster than our first solution! The reason for this comes from the fact that, during fitting, the parameters are changed very often to numerically calculate the derivatives of our function with respect to its parameters. Therefore lastX, lastY are reset most of the time.
You will see, however, that this function is much faster for plotting or for Monte-Carlo fitting, because these operations don’t call First as often as the Levenberg-Marquardt fitting algorithm.
Faster solution 3: using Derivatives
Even though we have just been disappointed by the method “using the last result”, we can improve this method and make fitting drastically faster.
The idea behind it is the following: The previous approach has failed because the parameters change too often during fitting. They are changed because, during fitting, the derivatives of the function with respect to its parameters are required at each data point. If you don’t define a procedure Derivatives in your function, these derivatives are calculated numerically, by slightly varying a parameter and calling your function again. This is the reason why the parameters change so often.
Once we have realised this, we know how to circumvent the problem: We must define the procedure Derivatives and calculate the function’s derivatives in respect to its parameters ourselves.
Now things are becoming complicated: How do I implement a fast, efficient way for calculating the derivatives?
Well, we could look for an analytical solution – but, again, this is not the subject of this TechNote.
For calculating the derivative of our function y(t) in respect to parameter ai numerically, we use the approximation:
dy(t)/dai = ( y(t, ai + e) – y(t, ai) ) / e (7)
where e is a small number. To make the integration of y(t, ai + e) fast, we now provide temporary storage not only for y(x, a1, a2, a3, a4) (such as we did in the above solution 2), but also for y(x, a1+ e, a2, a3, a4), y(x, a1, a2+ e, a3, a4), y(x, a1, a2, a3+ e, a4), and y(x,a1, a2, a3, a4 +e).
Here is the result:
function NonlinearDecay4;
const aStep = 0.01;
epsilon = 1e-8; {a small deviation for all params}
var lastX0, lastY0, lastX1, lastY1, lastX2, lastY2,
lastX3, lastY3, lastX4, lastY4;
function Integration(lastX, lastY, xVal, a1, a2, a3, a4);
{runs one integration}
var yVal, t, dt;
begin
if xVal >= lastX then begin
yVal := lastY; {start value}
t := lastX; {start time}
end
else begin
yVal := a4; {start value}
t := 0; {start time}
end;
dt := aStep; {step width}
while t < xVal do
begin
if t+dt < xVal then dt := xVal-t;
yVal := yVal + dt*(yVal*(a1 + yVal*(a2 + yVal*a3)));
t := t+dt;
end;
Integration := yVal;
end;
procedure First; {called when lastX, lastY are stale}
begin
lastX0 := 0; lastY0 := a[4]; lastX1 := 0; lastY1 := a[4];
lastX2 := 0; lastY2 := a[4]; lastX3 := 0; lastY3 := a[4];
lastX4 := 0; lastY4 := a[4]+epsilon;
end;
procedure Derivatives;
var yVal, yVal1;
begin
yVal := Integration(lastX0, lastY0, x, a[1], a[2], a[3], a[4]);
lastX0 := x; lastY0 := yVal;
yVal1 := Integration(lastX1, lastY1, x, a[1]+epsilon, a[2], a[3], a[4]);
lastX1 := x; lastY1 := yVal1;
dyda[1] := (yVal1-yVal)/epsilon;
yVal1 := Integration(lastX2, lastY2, x, a[1], a[2]+epsilon, a[3], a[4]);
lastX2 := x; lastY2 := yVal1;
dyda[2] := (yVal1-yVal)/epsilon;
yVal1 := Integration(lastX3, lastY3, x, a[1], a[2], a[3]+epsilon, a[4]);
lastX3 := x; lastY3 := yVal1;
dyda[3] := (yVal1-yVal)/epsilon;
yVal1 := Integration(lastX4, lastY4, x, a[1], a[2], a[3], a[4]+epsilon);
lastX4 := x; lastY4 := yVal1;
dyda[4] := (yVal1-yVal)/epsilon;
end;
begin
y := Integration(lastX0, lastY0, x, a[1], a[2], a[3], a[4]);
lastX0 := x;
lastY0 := y;
end;
Again, for simplicity, we don’t use Runge-Kutta here. Note that the integration is done in the function Integrate. lastX0, lastY0 are the temporary buffers for the unperturbed parameters, lastX1, lastY1, lastX2, etc. are the buffers for perturbed a[1]+epsilon, a[2]+epsilon, etc.
The function NonlinearDecay4 looks daunting. But it was worth it: Fitting now less than 1 second, which is more than an order of magnitue faster than our original solution.
To make NonlinearDecay4 even faster, we could implement Runge-Kutta in the function Integrate, but we leave this as an exercise for the interested reader.
There are many other ways to make your function faster, e.g. by optimising the (presently not optimised) step width in the constant “aStep”. You may also try to store more than one calculated y(t) for each data set (e.g. by doing a full integration in procedure First and filling arrays with the results, such that the function can directly access the results in the arrays). Often, you may also be able to optimize your models by having a good look at your equations and trying to simplify them.
A word about our model
In our model we assumed that the alcohol decreases with the law given in Eq. (4), i.e. the absorption rate is a sum of a linear and square term. A more realistic model might start from the assumption
dy/dt = k1 (1 – exp(-y/k2))
This equation is linear in y for small values of y, while it saturates to k1 for large values of y. In other words, it says that for large alcohol concentration, the liver is working as hard as it can to remove alcohol (the rate of absorption is limited by the liver capacity and independent of the alcohol concentration) while for small alcohol concentration, the liver has sufficient capacity to remove every alcohol molecule it can get hold of (the rate of absorption is proportional to the alcohol concentration).
We leave the implementation of the above equation as an exercise for the interested reader.
Coupled differential equations
Differential equations are highly social beings, therefore they often come in groups. Such groups are called coupled differential equations. Example:
dy1/dx = f1(x, y1, y2, a1 … aN)
dy2/dx = f2(x, y1, y2, a1… aN) (8)
Where y1 and/or y2 are measured values at points x, and a1 … aN are the parameters to be fitted. f1 and f2 are known functions.
To fit such a set of equations, you must define a function that integrates y1 and y2 from a starting point up to x and then returns y1(x) or y2(x), depending on which value you want to fit. In its most simple form, such a function could look as follows:
function DifferentialEquations;
const aStep = 0.01;
var t, dt, y1, y2;
begin
y1 := a[1]; {start value for y1}
y2 := a[2]; {start value for y2}
t := 0; {start time}
dt := aStep; {step width}
while t < x do
begin
if t+dt > x then dt := x-t;
y1 := y1 + dt*f1(t,y1,y2, a[1]... a[N])
y2 := y2 + dt*f2(t,y1,y2, a[1]... a[N])
t := t+dt;
end;
y := y2;
end;
In the above example, you must replace f1 and f2 with your actual functions.
Obviously, you can again make this function faster by using the techniques described above. Furthermore, if you have measured y1 as well as y2, you can fit both of them simultaneously using one of the methods described in the chapter on fitting multiple functions of the pro Fit user manual.
The following example describes a simple implementation of two coupled differential equations.
Example 2 – Rabbit Island


Rabbit Island is a tiny spot somewhere in the Atlantic ocean. In former days, it used to be called Grass Island because there was nothing but grass on it. Then, some fine day, a Viking ship visited it. On board of the Viking ship, there were ten rabbits, which were supposed to provide a festive meal on Eric the Red’s birthday.
While the ship was anchored at the island, the winds carried the smell of rich, green grass to the noses of the little rabbits, who seized the day, jumped over board and swam to the shore. The Vikings, unaware of their meal’s excursion, sailed off and were never seen again. The rabbits were left behind, happily munching the green treasures of their new home, Rabbit Island.
There are some exciting mathematics behind this story. Before digging into it, a word of warning: The mathematical model shown here was invented to illustrate how to solve coupled differential equations with pro Fit – it is not meant to be a sound scientific model of what is happening on Rabbit Island.
The mathematics of Grass Island
Before plunging into the complicated formulas describing daily life on Rabbit Island, let us have a brief look at the rules that describe life before the advent of the rabbits, i.e. life on Grass Island.
On Grass Island, there is only one single life form, the grass. The grass grows and dies. Let us call the amount of grass g. g varies in time according to the following equation:
dg/dt = gGrow·g – gDie·g2 (10)
The first term, “gGrow·g”, describes the rate of “natural” grass birth and death: The number of new grass seeds per time unit is proportional to the amount g of grass that is already there, and the number of grass plants dying of old age is also proportional to the present amount g of grass. The constant gGrow takes both these processes into account.
The second term, “- gDie·g2“, stands for the amount of grass dying because of overpopulation: The chance that a single grass plant dies because of overpopulation in a unit of time is proportional to the number of other grass plants around, hence the total number of grass plants dying because of overpopulation is proportional to the square of the number of existing grass plants, i.e. proportional to the square of g.
On Grass Island, before the advent of the rabbits, gGrow was 1, gDie was 0.01. To find the “steady state” solution of Eq. (10), i.e. the value of g that was finally reached on Grass Island, we simply set dg/dt to 0 and calculate g = 100. (g is in grass units, where one grass unit equals 31415.926 grass plants.)
The time behaviour of Eq. (10) is rather boring. We won’t look at it in detail. We simply assume that at the time the rabbits arrived on the island, the grass was in its steady state, i.e. g was 100.
The mathematics of Rabbit Island
As everyone knows, rabbits eat grass. Hence, after the arrival of the little rodents, Eq. (10) must be corrected with a term describing the amount of grass eaten. This amount is proportional to the probability that a rabbit finds and eats a grass plant, i.e. proportional to the number of rabbits and grass plants. Hence, we must introduce a term proportional to g*r, where r is the number of rabbits:
dg/dt = gGrow·g – gDie·g2 – gEaten·r·g (11)
Rabbits have babies and rabbits may die of old age or starvation (there are no predators or diseases on Rabbit Island). We assume that the number of rabbits being born is proportional to the food resources that are available as well as to the number of rabbits already living, i.e. proportional to g·r. The number of rabbits dying of old age is simply proportional to the number of rabbits that are on the island. Hence, the time derivative of the number of rabbits r follows the following law:
dr/dt = rGrow·r·g – rDie·r (12)
(To further refine this, we could add a “starvation” term depending on r/g, the ratio of rabbits to available grass. But for simplicity, we stick with Eq. (12).)
Equations (11) and (12) are two coupled differential equations, such as they are shown in Eq. (8). Hence, we can solve them as described above. The following is a simple pro Fit function doing this job for us:
function RabbitIsland;
inputs
a[1]:=0.01,active, 'r grow';
a[2]:=0.003,active, 'r die';
a[3]:=1,active, 'g grow';
a[4]:=0.01,active, 'g die';
a[5]:=0.01,active, 'g eaten';
a[6]:=10,active, 'r0';
a[7]:=100,active, 'g0';
a[8]:=0.1,constant,'step';
outputs
y[0]:=0, 'rabbits';
y[1]:=0, 'grass';
var t, dt;
r, g;
procedure Initialize; {called once after compilation}
begin
SetFunctionProperties(preview allOutputValues); // preview should also show the grass
end;
begin
r := a[6]; {start value for rabbits}
g := a[7]; {start value for grass}
t := 0; {start time}
dt := a[8]; {step width}
while t < x do
begin
if t+dt > x then dt := x-t;
r := r + dt*(a[1]*r*g - a[2]*r);
g := g + dt*(a[3]*g - a[4]*g*g - a[5]*r*g);
if r < 0 then r := 0; {all died}
if g < 0 then g := 0; {all died}
t := t+dt;
end;
y[0] := r;
y[1] := g;
end;
In this example, a[1] is rGrow, a[2] is rDie, a[3] is gGrow, a[4] is gDie, a[5] is gEaten, a[6] is r0 (the number of rabbits at the beginning,10), a[7] is g0 (the amount of grass at the beginning, 100 grass units).
In our example function we use a primitive one step integration algorithm. The step width for integration is given in a[8]. If we have to use our function often, we could use one of the more efficient algorithms (“Using the last result”, “Runge-Kutta”, “Using derivatives”) that are described above. For an example of the implementation of the algorithm “Using the last result”, have a look at the file “Rabbit Island Functions” that comes with this TechNote.
The development of r and g in time depends strongly on the values used for parameters a[1] to a[7]. For the default parameters listed above, we get the following result:
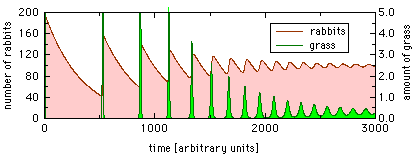
As you can see, the number of rabbits (the upper curve) immediately rises from its starting value 10 to about 200. This comes from the fact that the rabbits encounter an island that is covered with grass. During this drastic, initially exponential, increase of the rabbits population, the amount of grass drops to nearly 0 and the rabbits run out of grass. They start starving because there’s nearly no food available until time 500. Then the number of rabbits has fallen so low that the grass can recover, which accounts for the grass peak right after time 500. This provides food for the rabbits, which multiply another time until they have again eaten nearly all grass, so they start starving again. This process continues, slowly loosing in amplitude. Approximately at time 2000 the system starts settling at an average value of 100 rabbits and g = 0.3.
It is quite interesting to vary the parameters and watch their influence on r and g. For instance, when you increase the parameter rDie (the natural death rate of the rabbits), you will see that the oscillations become faster – which leads to a more realistic behaviour because the rabbits die faster in the absence of food. Decreasing rGrow (the growth rate of the rabbits) decreases the amplitude of the oscillations and the time until the system finds its steady state. If the growth rate rGrow is sufficiently low, the oscillations disappear completely.
As explained above, the Vikings left the island after the rabbits escaped. No one else has found Rabbit Island yet. Some think the island is a myth, others believe that it has sunk into the depth of the ocean. Therefore, we don’t know really how the rabbits fared in their new home, and we don’t have any experimental data on r(t). If we had, we could fit our function to test our mathematical model and find the values of its parameters.
Where to go from here?
Even though its mathematics are simple, the behaviour of Rabbit Island is quite complex. Our model is certainly not sufficient for explaining reality.
For instance, we ignore that young rabbits can have no babies, while old rabbits die more often of old age. To improve our model, we could therefore set up separate differential equations for rYoung and rOld, where rYoung is the number of young rabbits, rOldthe number of old ones. We would then end up with three coupled differential equations. To refine our model, we should also take into account that rabbit droppings are a good grass fertiliser and add an equation for the amount of rabbit droppings on Rabbit Island.
Furthermore, there is an unconfirmed rumour that the rabbits were not the only ones escaping from the Viking ship to the island: Some dogs left it as well. And dogs eat rabbits! Another differential equation.
Differential equations similar to Eq. (11) and (12) can be used for the description of many other systems with interacting “populations” of various “species”, such as the number of molecules in ground state and excited states of a dye laser, the electron and hole densities in different parts of a semiconductor, the water molecules in different sections of a volume of ground, etc.
Similar (or somewhat more complicated) models might also approximate larger systems. Consider setting r to the number of people on this world, g to the amount of natural (renewable) resources. Think about it!